The idea of identifying new and alternative indications for already approved/pioneering, shelved/failed compounds or even drug candidates that are under development is termed drug repurposing or repositioning. It remains a viable alternative to conventional de novo drug target identification and development. This is an attractive means with too little attention to maximize return on preclinical and clinical investments of pharmaceutical companies that were originally intended for different target populations. Repurposing of already existing and ‘derisked’ compounds as candidates for rare and untreated diseases hopes to overcome the myriad of problems faced by pharmaceutical companies by minimizing escalating costs for development, investment in clinical trials, limiting stagnant productivity, mitigate attrition/failure rates and shorten the timeline for development and thus aims at encouraging more investment for finding cures for untreated/neglected diseases. The timeline for development for repositioned drugs can be 30–60% lower than those for de novo compounds. The overall cost of development can be reduced by as much as 60% of those for new chemical entities since existing drugs have been subjected to regulatory review, and undergone safety profile trials previously. The probability of success for a repurposed drug is significantly higher than a fully new chemical or biological entity. Rescuing previously shelved candidates could also lead to the recovery of R&D expenses, if successful, the return is multiple. In 2015, repositioned drugs were estimated to account for 25% of the revenue of the pharmaceutical industry and it is estimated that it could be significantly more.
With the increasing largescale, publicly available comprehensive data on existing drugs, diseases, omics, biomedicine and clinical trials. High-throughput genomics and bioinformatics technologies along with Artificial Intelligence (AI) allow drug repurposing to facilitate and optimize drug discovery and development. AI allows effective integration of the different data and has the potential to identify relationships and/or derive connections that are highly unlikely to be identified by a researcher. Current computational and bioinformatics tools provide significant assistance to researchers, but much of the success of drug repositioning using such tools is determined by the ability of scientists to understand and use those tools and interpret the results generated. Applying AI to drug repositioning greatly increases the efficiency of the process and interpretation of the results thereby significantly reducing the time and cost of drug development and increasing predictability.
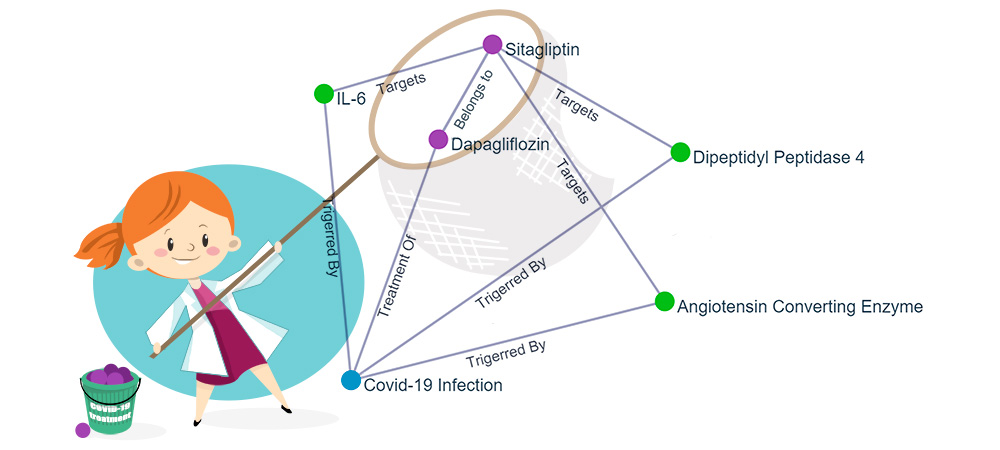
Keystonemab exploits specialized crawlers to gather relevant data from large scientific repositories containing millions of scientific publications and public domain life science websites. Information on drugs and diseases is then derived and later organized using the ‘Natural Language Processing’ algorithm where keywords and semantic relationships are extracted to establish our knowledge graph. Keystonemab contains comprehensive information on 95% of drugs and over a million relationships among drugs, diseases, drug target proteins, biomarkers and signalling pathways. AI allows for a comprehensive understanding of the context to establish parameter links between the publications. The relationship gives an understanding and guides scientists and key decision-makers towards applying their drugs for other disease indications and potentially reposition their drug.
Keystonemab’s AI platform also identifies other active ingredients influencing the similar signalling pathways, drug target proteins and biomarkers and thus allows a multiple, synergistic approach for an indication, which we refer to as the ‘Fishing Net Effect’ or ‘Network Effect’.